LC/MS/MS Method Package for Primary Metabolites Ver. 2
Описание
Ready-to-Use Analytical Conditions
Shimadzu Method Packages deliver conditions for efficient and simultaneous multi-component analysis. They enable the user to quickly and easily implement complex methods without costly and laborious method development by providing sample preparation protocols, LC separation conditions, and MS acquisition parameters.
Two Selectable Analysis Methods
Customers can select an appropriate analysis method from the two methods; analysis using an ion pairing reagent (55 components) and analysis using a PFPP column (97 components) according to the instrument conditions or metabolites to be analyzed.
Supporting LCMS-8040/8045/8050/8060
LC/MS/MS Method Package for Primary Metabolites Ver. 2 supports LCMS-8040/8045/8050/8060, enables high-sensitivity simultaneous multi-component analyses of primary metabolites.
Complete from Sample Preparation to Analysis
Protocols are included for the preparation of extracts from biological tissue. Saving the user time and money, even laboratories unfamiliar with extraction can follow prescribed steps for LC/MS/MS sample preparation.
Normalization of Multiple Sample Results
Due to mass differences between biological tissue samples, normalization of results must be performed when multiple samples are analyzed. This method package includes optimized analytical conditions for two internal standards to permit normalization across multiple samples.
Improved Data Analysis Functionality in Method Package
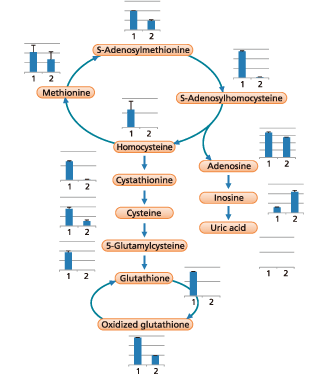
A wide variety of new data analysis functions have been added to the LC/MS/MS Method Package for Primary Metabolites. Functions such as automatically pasting data on metabolic maps, using a bar graph to compare changes in compound quantities, or graphing the changes as a function of time, for example, can be performed using simple operations. It also enables correlation analysis, volcano plots, displaying data on metabolic maps for target compounds, and other functionality. Consequently, it can significantly reduce the amount of work required for bottleneck processes such as data analysis and visualization, so that all operations from measurement to data analysis can be performed smoothly.
Data Analysis Tools Used in Method Package
- Volcano Plot
A tool that combines a t-test (statistically significant difference) and a fold analysis (Example: Difference in mean value such as 2 times or 1/2) to visualize the differences between the 2 groups. The Volcano Plot gadget developed by Shimadzu is included in the package. - VANTED
Tool maintained at University of Konstanz, Germany, for visualization and analysis of networks across different data sets. (GARUDA support was developed at Monash University) - iPath
Data analysis tool developed by the European Molecular Biology Laboratory that can be used for visualization of diverse metabolic pathways or data mapping and customization. - Cytoscape
Bioinformatics tool developed by the Cytoscape Consortium, used to visualize metabolic pathways, to integrate gene expression profiles with related data, and so on. It is especially useful for analyzing networks and visualizing correlations.
Index of compounds
| List of compounds for ion pair method | |
|---|---|
| Glycolytic system | Nucleotides |
| 2,3-Bisphosphoglyceric acid 3-Phosphoglyceric acid (2-Phosphoglyceric acid) Dihydroxyacetone phosphate Fructose 1,6-bisphosphate Glucose 1-phosphate Glucose 6-phosphate Glycerol 3-phosphate Phosphoenolpyruvic acid Pyruvic acid |
Adenosine 3′,5′-cyclic monophosphate Adenosine diphosphate Adenosine monophosphate Adenosine triphosphate Cytidine diphosphate Cytidine monophosphate Cytidine triphosphate Guanosine 3′,5′-cyclic monophosphate Guanosine diphosphate Guanosine monophosphate Guanosine triphosphate Thymidine diphosphate Thymidine monophosphate Thymidine triphosphate Uridine diphosphate Uridine monophosphate Uridine triphosphate |
| Co-enzyme | Pentose-phosphate pathway |
| NAD NADH NADP NADPH |
|
| 6-Phosphogluconic acid Erythrose 4-phosphate Ribose 5-phosphate Ribulose 5-phosphate Sedoheptulose 7-phosphate |
|
| TCA cycle | Internal STDs |
| Acetyl coenzyme A Succinyl coenzyme A |
2-Morpholinoethanesulfonic acid Methionine sulfone |
| Amino acids | |
| Alanine Arginine Asparagine Aspartic acid Cysteine Glutamic acid Glutamine Glycine Histidine Lysine Methionine Phenylalanine Serine Threonine Tryptophan Tyrosine |
|
| List of compounds for PFPP column method | |
|---|---|
| Glycolytic system | Organic acids |
| Lactic acid Pyruvic acid |
>4-Aminobutyric acid Adenylsuccinic acid Argininosuccinic acid Cholic acid Creatine Nicotinic acid Ophthalmic acid Orotic acid Pantothenic acid Taurocholic acid Uric acid |
| TCA cycle | |
| 2-Ketoglutaric acid Aconitic acid Citric acid Fumaric acid Isocitric acid Malic acid Succinic acid |
|
| Amino acids | Nucleosides and Nucleotides |
| 4-Hydroxyproline Alanine Arginine Asparagine Aspartic acid Asymmetric dimethylarginine Citrulline Cystine Dimethylglycine Glutamic acid Glutamine Glycine Histidine Homocystine Isoleucine Leucine Lysine Methionine sulfoxide Ornitine Phenylalanine Proline Serine Symmetric dimethylarginine Threonine Tryptophan Tyrosine Valine |
Adenine Cytosine Guanine Thymine Uracil Xanthine Adenosine Cytidine Guanosine Inosine Thymidine Uridine Adenosine 3′,5′-cyclic monophosphate Adenosine monophosphate Cytidine 3′,5′-cyclic monophosphate Cytidine monophosphate Guanosine 3′,5′-cyclic monophosphate Guanosine monophosphate Thymidine monophosphate |
| Co-enzymes | |
| FAD FMN NAD |
|
| Methylation and Transsulfuration cycle | Others |
| Cystathionine Cysteine Homocysteine Methionine |
2-Aminobutyric acid Acetylcarnitine Acetylcholine Allantoin Carnitine Carnosine Choline Citicoline Creatinine Cysteamine Dopa Dopamine Epinephrine Histamine Hypoxanthine Kynurenine Niacinamide Norepinephrine Serotonin |
| 5-Glutamylcysteine Glutathione Oxidized glutathione S-Adenosylhomocysteine S-Adenosylmethionine |
|
| Internal STDs | |
| 2-Morpholinoethanesulfonic acid Methionine sulfone |
|
* In this method package, customer can select an appropriate analysis method from the two methods using ion pairing reagent (57 components) and PFPP column (97 components) according to the instrument conditions or metabolites to be analyzed.*
The analysis method using ion-pairing reagents is recommended for fluctuation analysis of primary metabolites related to major metabolic pathways such as glycolysis, the pentose phosphate cycle, diphosphate and triphosphate nucleotides. While, the analysis method using PFPP columns is recommended for analysis of amino acids, organic acids (including the TCA circuit), the Methylation cycle and the Urea cycle.
Remarks and Precautions:
For Research Use Only. Not for Use in Diagnostic Procedures.
LabSolutions LCMS Ver.5.93 or later is required.
Shimadzu makes no warranty regarding the accuracy of information included in the database or the usefulness of information obtained from using the database.
It is the user’s responsibility to adopt appropriate quality control tests using standard samples to confirm qualitative and quantitative information obtained with this method package.
LabSolutions and LCMS are trademarks of Shimadzu Corporation.
GARUDA is a trademark of The Systems Biology Institute.